Dear David,
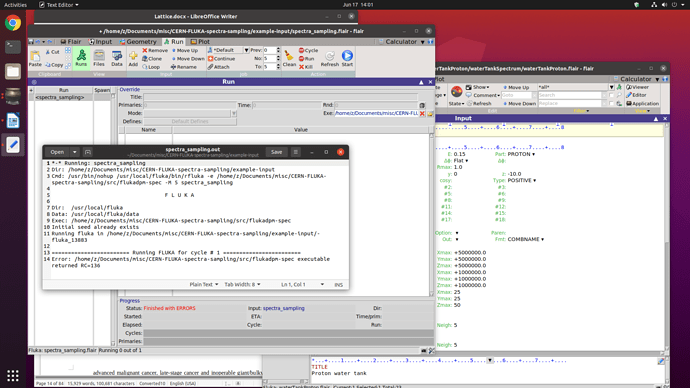
I also try to run the example input provided in the Sampling from energy spectra, the result came out the same, returning RC=136
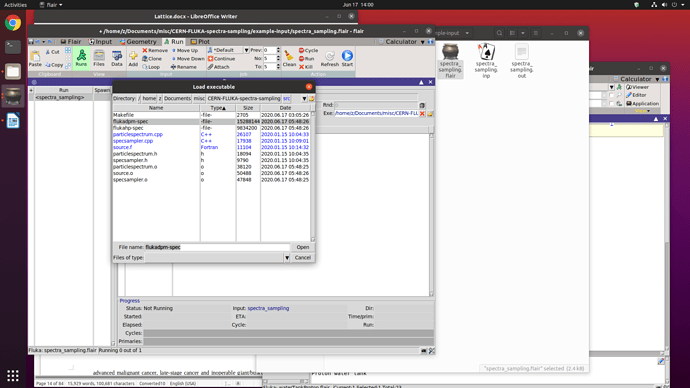
Here is a screenshot where I override the executable file
and the error message in the .out file.
Thank you!
Z.