Dear FLUKA and FLAIR users,
We are using im my team a CT device that does not generate standardized DICOM images in CT number/HU unit. I want to import the images to run a simulation with a poly-energetic source (between 10 and 118keV photons).
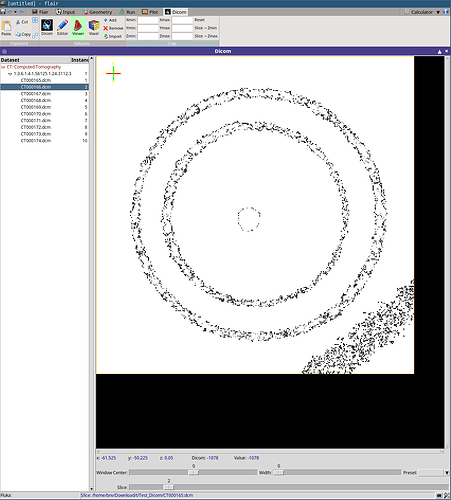
I am having an issue with DICOM images I have generated myself from a python code (and pydicom).
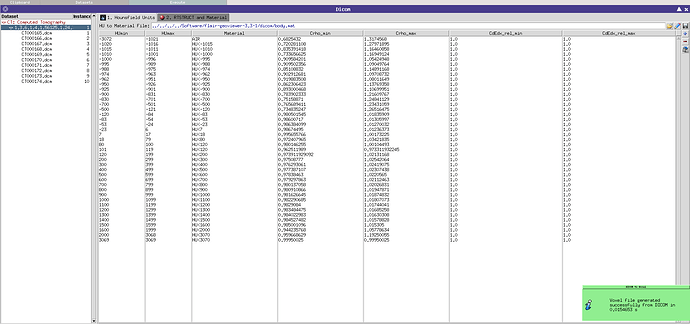
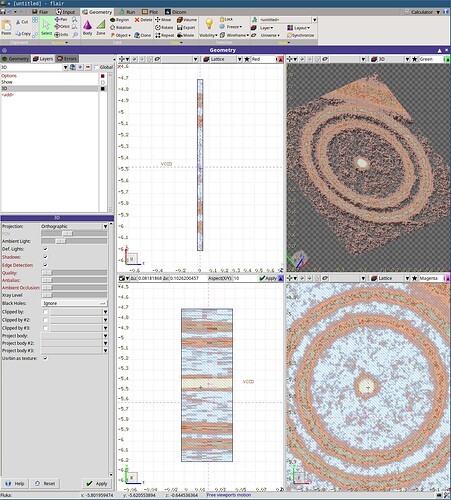
In the DICOM tab, I am importing my dataset but I can not see it with the DICOM viewer. However, I can generate a .vxl file with the “Voxel” tool and a successful output message (cf screenshot). But then in the geometry tab, the material is assigned to “error”.
The only log I have comes from my unix console where this message is displayed:
e> ERROR: Invalid voxel file. Data size do not match
VOXEL Title: Chambre RaySafe,
nx=300 ny=300 nz=1
dx=0.005 dy=0.005 dz=0.005
Dx=1.5 Dy=1.5 Dz=0.005
nregions=183 nvoxels=459
ERROR wrong data length=600 expected=180000
ERROR reading voxel file “test.vxl”
Do you know what is this issue? Is there a way to get more log information? Is there another way to generate a .vxl file
Test_Dicom.zip (1.3 MB)
?
Thank you for your help,
test.vxl (16.9 KB)
Mars_PMMA_Center.flair (7.0 KB)
histogram_114kVp_HVL_5.48mmAL_Sampling_0.8keV.txt (1.6 KB)
source_newgen.f (18.8 KB)