Dear FLUKA Experts,
For radiation protection studies, I am working on simulating radiation from isotopic sources based on the activity of their constituent nuclides. As I understand, this typically requires the semi-analogue mode.
Since a source may contain hundreds of isotopes, using individual HIGH-PROPE cards and #define conditions becomes difficult due to computational limits on isotope count and computational load. To work around this, I have written a custom source.f routine that reads an input file and assigns isotopic properties accordingly. While it functions as intended, I have come to realize, that my approach may not align with how semi-analogue mode actually handles decay chains, thanks to this insightful post.
Based on the helpful explanation in the referred post, I believe I made at least 2 mistakes:
- When both parent and daughter nuclides are included, semi-analogue mode simulates the full decay chain of each of them, which can overestimate daughter radiation. In shielding studies this is often conservatively acceptable, though it does not represent the true activity distribution.
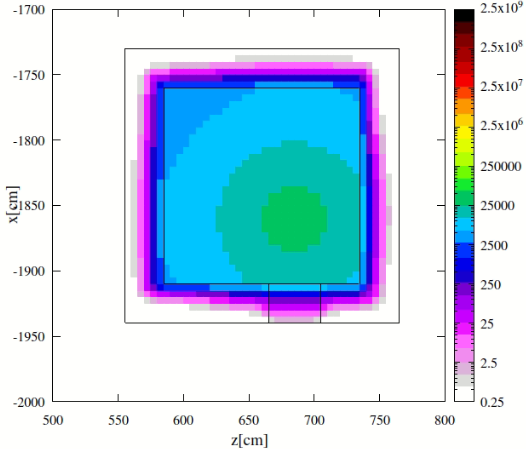
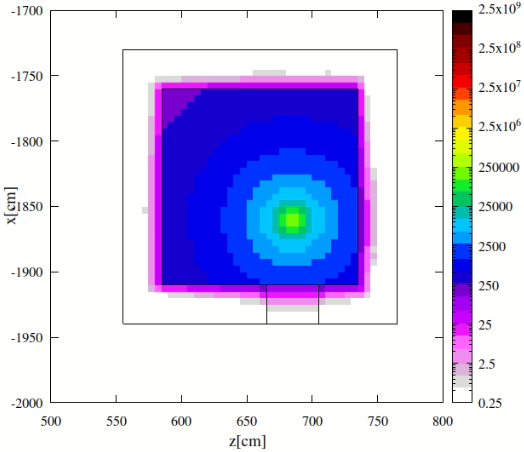
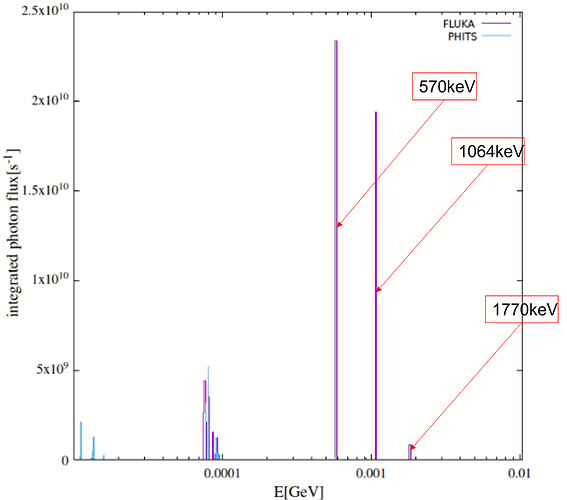
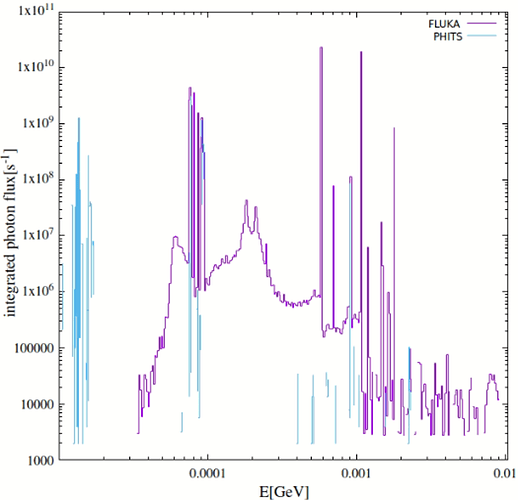
- More importantly, the sampling of nuclides in this mode can lead to inaccurate daughter radiation intensities, as theoretically outlined in the post. This became apparent in my At‑211 case. At‑211 decays mainly via alpha emission with weak gamma yield, while its long-lived daughter Bi‑207 emits strong gamma rays around 1063 keV. In practice, Bi‑207 activity remains low in an At‑211 source, making it a relatively weak source. However, my FLUKA simulations in semi-analogue mode suggested otherwise—a discrepancy that I suspect may stem from the sampling mechanism (I have compared with PHITS results, which I can share if helpful).
While semi-analogue mode appears to provide a “time-averaged” response, my goal is to obtain a “prompt decay” response, scoring only radiation from the nuclides explicitly specified in the source.
My main question remains: Is there a recommended way to exclude daughter nuclide contributions?
I have tried TIME-CUT and TCQUENCH with a t=0 threshold, but parent decays do not seem to take place at t=0. I also tested LTRACK , but only saw a value of 2. If daughters were labeled with higher LTRACK values, distinguishing them would be straightforward—though that does not appear to be the case.
Could LTRACK still be useful in this context? If so, I would appreciate any hints on what I might have missed. If not, I would be grateful for suggestions on how to write a user routine to achieve this filtering.
iso_source.f (12.2 KB)
mgdraw.f (14.9 KB)
model-v5.flair (12.3 KB)
nuc_input(Bi_5hAt).dat (40 Bytes)