Dear @horvathd
I successfully calculated the dose distribution. Now the problem is, I found that the RTViwer module has been removed in the new version.
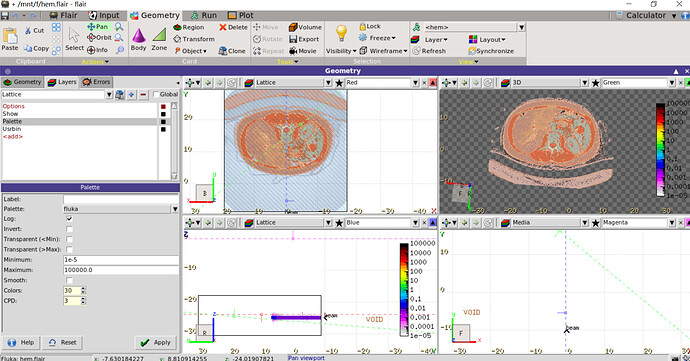
Is there a way to superimpose the dose distribution on the CT image like the flair manual? I tried to superimpose the dose on the CT image in the geomtry interface, but it failed.
Thank you
Ye
Dear @ye.jiaquan
You have the following options:
- In the palette tick the Transparent (< Min) so as all the white color to become transparent. Of course you need to adjust the minimum value accordingly
- You can use the transparency option in the “Usrbin” to blend the colors
- If you have an RTDOSE file equivalent to the USRBIN you can use the RTViewer (if you don’t have an RTDOSE otherwise the rtviewer is not activated). However in the Dicom tab you can convert your USRBIN to RTDOSE and use the RTViewer afterwards
Dear @vasilis
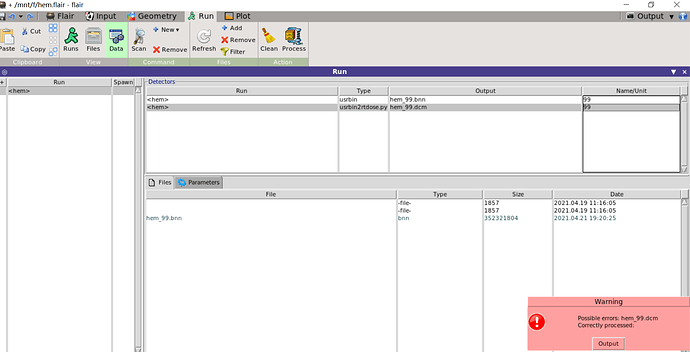
I try to convert USRBIN to RTDOSE by usrbin2rtdose.py,unfortunately the error was reported
e> ERROR: RTDOSE file not specified
e> ERROR: cannot access file ‘hem_99.dcm’
Maybe the run is still going on?
Error processing: hem_99.dcm
Ended: 2021.04.21 19:48:24
Warning
Possible errors: hem_99.dcm
Correctly processed:
Thak you for your help!
Ye
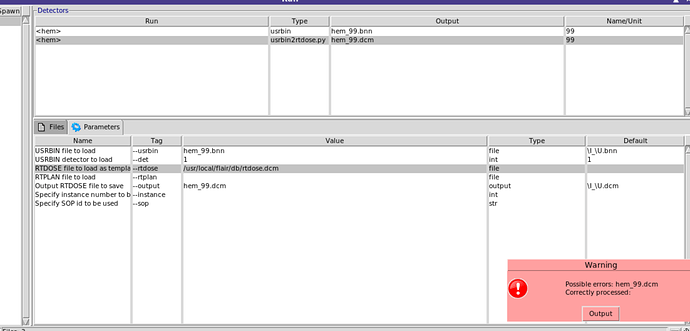
When you click on the tab Parameters you need to provide the information there
Dear @vasilis
I don’t have RTDOSE and RTPLAN file, are they necessary to convert usrbin to RTDOSE?
Also, I tried the methods mentioned in this topic
error is reported
e> Usrbin to RTDOSE conversion:
Usrbin: /mnt/f/medical data/06-05-2000-522-26569/4.000000-RT Dose - fx1hetero-9.5.1/hem_99.bnn
RTDOSE: /mnt/f/medical data/06-05-2000-522-26569/4.000000-RT Dose - fx1hetero-9.5.1/000000.dcm
e> ERROR: Cannot open RTDOSE ‘/mnt/f/medical data/06-05-2000-522-26569/4.000000-RT Dose - fx1hetero-9.5.1/000000.dcm’
‘NoneType’ object has no attribute ‘read_file’
Thank you
Ye
Dear @ye.jiaquan
Normally the rtplan if it is not supplied it should use 1.0 as normalization. However in the distributed program it was exiting.
Could you please test the modified usrbin2rtdose.zip (4.5 KB)
It contains also the rtdose.dcm template file to be used in case none is supplied. It expects to find the file inside the db subdirectory of flair
Dear @vasilis
I try to use the new usrbin2rtdose file and load the rtdose.dcm template file,unfortunately the error is reported
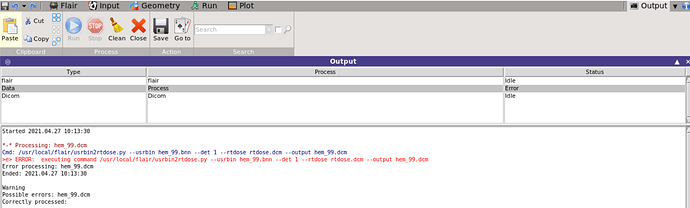
- Processing: hem_99.dcm
Cmd: /usr/local/flair/usrbin2rtdose.py --usrbin hem_99.bnn --det 1 --rtdose /usr/local/flair/db/rtdose.dcm --output hem_99.dcm
e> ERROR: Cannot open RTDOSE ‘/usr/local/flair/db/rtdose.dcm’
‘NoneType’ object has no attribute ‘read_file’
Usrbin to RTDOSE conversion:
Usrbin: hem_99.bnn
RTDOSE: /usr/local/flair/db/rtdose.dcm
e> ERROR: cannot access file ‘hem_99.dcm’
Maybe the run is still going on?
Error processing: hem_99.dcm
Ended: 2021.04.24 18:46:55
Warning
Possible errors: hem_99.dcm
Correctly processed:
Dear @ye.jiaquan
The “read_file” error is reported from the line
pydicom.read_file(rtdose_file, force=True)
It seems to me that you are missing the pydicom package on your system.
Dear @vasilis
I install flair through the flair repository.It seems that pydicom has been successfully installed.
Thank you
Ye
Which linux system you have? In my ubuntu 20.10 is called python3-pydicom
Dear Ye,
for Ubuntu 18.04 or 20.04 (used for WSL) you need to install the python3-dicom package.
Cheers,
David
Dear @horvathd @vasilis
I use Ubuntu 18.04 and python 2.7. think python3-dicom has been successfully installed.
Thank you
Ye
Apparently the ubuntu 18.04 is still using the old dicom library that was named dicom instead of the new pydicom.
Please try the following script that should correct the problemusrbin2rtdose.txt (11.2 KB)
(rename it to: usrbin2rtdose.py instead of .txt)
Dear @ye.jiaquan could you please send your usrbin file so I can test on my computer
Dear @vasilis
Of course, here is the usrbin file.
I don’t know what went wrong, but on my computer the conversion works ok. Please find attached the generated dicom filehem_99.dcm.zip (41.5 KB)
I will try on Ubuntu 18.04 to verify if there is something else that generates the problem
@ye.jiaquan The error that was displayed was wrong. The rtdose dicom file normally should have been generated, simply flair was not detecting it.
Until the next release that is fixing the problem you can run the usrbin2rtdose.py from the command line